The Zeiss ApoTome is an optical instrument which can be placed in the light path of a traditional epifluorescence microscope to reduce out-of-focus light and enhance optional sectioning. This is achieved by illuminating the specimen through a rapidly tilting a glass plate with a grid on it and analyzing the resulting image in real time to create an image which maximally excites and captures light at a single optical section (calculated from at least 3 raw images acquired at different tilt angles).


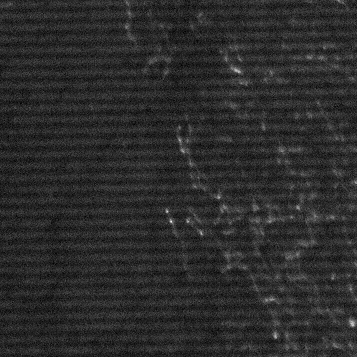
A poorly-calibrated ApoTome will produce imaging artifacts which appear as horizontal lines on the final image (in addition to increased collection of out-of-focus light). The ApoTome must be calibrated separately for every filter cube / lens combination. An ApoTome on a microscope with 4 filter cubes and 5 lenses must be calibrated 20 times. Calibration is a two-step process, requiring phase calibration and a grid calibration. See Imaging Associates - ApoTome Calibration Guide.
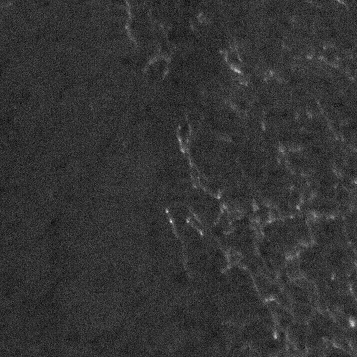
Although ApoTome artifact lines can be reduced by improving calibration, images with these artifacts can be improved with ImageJ. Since the artifacts are typically horizontal lines with a regular distance between them, they are ideally isolated (and eliminated) in the frequency domain. The conversion of linear data into a frequency domain can be accomplished using the Fast Fourier transform (FFT), and this has special uses for image processing when a 2D FFT is used. In our case we will convert the artifact-laden image to the frequency domain, identify the frequencies underlying the artifact, and perform an inverse FFT (iFFT) to create an original image with reduced artifacts.
Manual Reduction of ApoTome Artifacts
- Open an image with ApoTome artifacts
- Create an FFT (Process > FFT > FFT)
- Select black color (double click the dropper tool)
- Identify apotome artifact hot spots
- Hot spots appear as bright points of light (excluding the center point)
- Apotome artifact hot spits will be symmetrical on both sides of the center point
- If apotome lines are horizontal, the artifacts will be bright points along the horizontal mid-line of the image vertically offset from the center.
- If the apotome is rotationally misaligned, it will deviate from midline by the same angle of its misalignment.
- Most FFTs from apotome images have 2 hot spots, but its possible more than 2 are needed.
- Select the hot spots with the elipse tool
- Fill each hot spot with dark (CTRL+F)
- Create an inverse FFT (Process > FFT > Inverse FFT)
Automatic Reduction of ApoTome Artifacts
Manual reduction of ApoTome artifacts using the above method is labor-intense. If you have a stack with a large number of images, this process can be applied to every slice of the stack using the following ImageJ macro. No user interaction is required.
function blackout_FFT_for_apotome(){
/* Draw ovals around trouble frequencies of an apotome image FFT, then run IFFT.
* Position settings may need to be tweaked for your individual microscope.
*/
getDimensions(width, height, channels, slices, frames);
ovalVertOffset = 178;
ovalWidth = 120;
ovalHeight = 30;
ovalLeft = width/2-ovalWidth/2;
ovalTop = height/2-ovalHeight/2;
setForegroundColor(0, 0, 0);
makeOval(ovalLeft, ovalTop-ovalVertOffset, ovalWidth, ovalHeight);
run("Fill", "slice");
makeOval(ovalLeft, ovalTop+ovalVertOffset, ovalWidth, ovalHeight);
run("Fill", "slice");
run("Select None");
run("Inverse FFT");
}
function fix_apotome_artifacts_in_stack(){
/* Run this on a stack to remove apotome artifacts.
* This creates a second stack of corrected images.
*/
stackName = getTitle();
for (i=1; i<=nSlices; i++){
setSlice(i);
sliceName=getInfo("slice.label");
// create the FFT
run("FFT");
rename("FFT_"+sliceName);
blackout_FFT_for_apotome();
rename("IFFT_"+sliceName);
// delete the FFT
selectWindow("FFT_"+sliceName);
close();
// re-select the original window before looping
selectWindow(stackName);
}
run("Images to Stack", "name=Stack title=[] use");
}
fix_apotome_artifacts_in_stack();